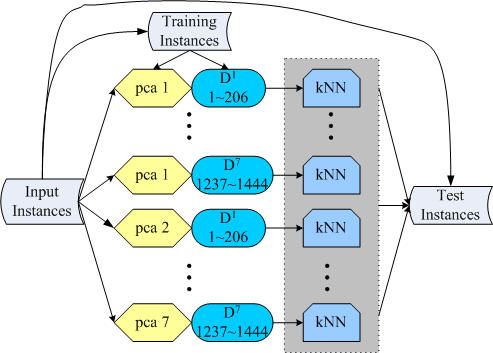
The flowchart of the
ensemble system for the drug-target prediction. |
Background: Drug-target interaction is key in drug
discovery, especially in the design of new leader
compound. However, the work to find a new leader
compound for a specific target is complicated and hard,
and it always lead to many mistakes. Therefore
computational techniques are commonly adopted in drug
design, which can save time and costs in a significant
extent.
Results: To address the issue, a new prediction system is proposed in
this work to identify drug-target interaction. First,
drug-target pairs are encoded with a fragment technique
and the adopted software "PaDEL-Descriptor". The
fragment technique is for encoding target proteins,
which divides each protein sequence into several
fragments in order and encodes each fragment with
several physiochemical properties of amino acids. The
software "PaDEL-Descriptor" creates encoding vectors for
drug molecules. Second, the dataset of drug-target pairs
is resampled and several overlapped subsets are
obtained, which input into kNN (k-Nearest Neighbor)
classifier to build an ensemble system.
Conclusion: Experimental results on the drug-target dataset showed that
our method performs better and runs faster than the
state-of-the-art predictors. |